- Home
- Careers
- Careers at Peter Mac
- Research careers
- Peter Mac Postdoc Society
- Bench and Beyond: Tales from Peter Mac Postdocs
Bench and Beyond: Tales from Peter Mac Postdocs
Bench and Beyond is a space where postdocs from diverse scientific and cultural background share stories about their journey into science, the focus of their research, the highs and lows they encounter and provide insights into their lives beyond work. Our goal is to highlight the research of Peter Mac postdocs, offering an authentic portrayal that resonates with aspiring scientists.
Dr Anna Trigos is a Postdoctoral Researcher in computational biology, bioinformatics and prostate cancer. Her expertise includes gene expression, genomics, gene networks and tissue microscopy image analysis. She won the Joseph Sambrook Prize for Research Excellence and the Postgraduate Research Medal in 2019.

The complexity of cancer biology has always amazed me. Regardless of how much you learn, discover or understand, it always feels like just the beginning, like nature has put up an impenetrable wall that the human mind could never break through.
My first research project as an undergraduate student in biology was looking at how one tiny molecule was controlled by another, and the role this played in nerve regeneration. I remember spending hours trying to understand how all the pieces of the puzzle fit together and why this molecule, among the hundreds of thousands of others, was special. At some point, however, I realized that it was impossible – it was just too complex for a human brain to remember all 20,000 gene names and all the pathways in which they are involved (although, I must say, I have met people who seem to have come close to this). I didn’t know it at the time, but it was at that point I decided to become a computational biologist, although I could barely write 3 lines of code!
As a cancer researcher in computational biology and bioinformatics, data (not pipettes!) are our bread and butter. People think of different things when you say data – “numbers,” “digits,” “storage” – but to me, it represents an undiscovered world of networks and patterns. One small change in one corner of a complex network could have a cascade effect and disrupt the entire system, transforming a normal cell to a cancer cell. Or perhaps random changes in gene activity or in the cells that aid tumor growth are part of a consistent – albeit complicated – pattern. This is the common ground of all the projects I have worked on – I look for ‘patterns,’ for ‘networks.’ In other words, I try to define the ‘rules’ that govern a system.
In my first computational project we wanted to explore whether cancer cells no longer relied on being part of an organ and started resembling single-cell organisms, such as bacteria. To answer this question, we were using data at the level of the activity of genes – that is, looking at what genes are switched on or off in cancers at any point in time. In practice this meant that we were looking for a ‘pattern’ among 20,000 genes and 10,000 samples (so 2x108 data points), where we didn’t know what the pattern was or how we could find it. In the end we uncovered an underlying architecture of biological networks, where key interactions were broken down by mutations. With this, we had added a novel layer to our understanding of the fundamental biology of cancer.
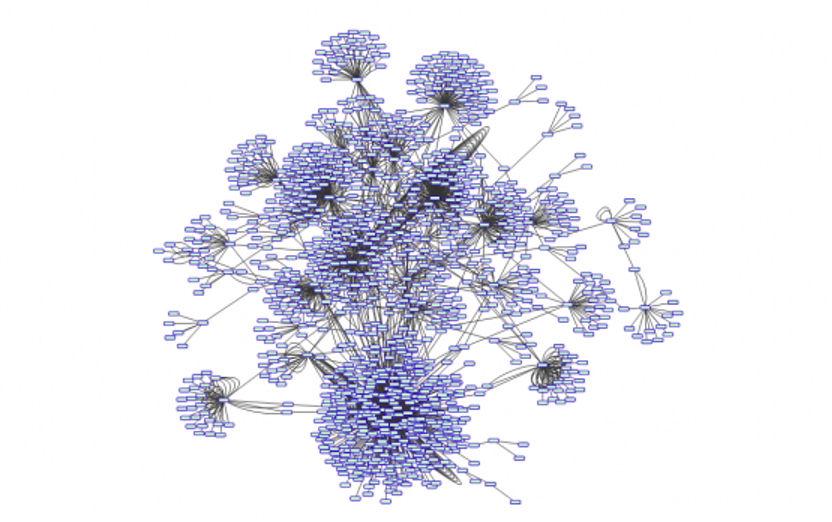
Figure legend: Networks of connections between genes involved in cancer, such as this one, often look like flowers. A coincidence or a hidden pattern?
My latest project is largely translational, where we are trying to identify prostate cancer patients likely to respond to immunotherapy. For this, we are using an exciting new type of data and analysis – spatial analysis of tissue microscopy images. Here too the computational aim is to find patterns, but of a different kind – that of how immune cells interact with tumor cells. Tissues, unfortunately, are messy, and my question here is less “what is the pattern” but rather “what can we measure and how can we do it?” They say that an image is worth a thousand words, but unfortunately, they do not give you p-values or data distributions that you could add to a publication.
We realized very quickly that we needed to build new tools to interpret this data. I am now integrating quantitative methods borrowed from the fields of geography and ecology to uncover and quantify hidden patterns without relying on the human eye. Up to now we have found that particular spatial patterns of immune cells are associated with either immunosuppression or tumor immune recognition, and we are interested in understanding how they link to disease progression, metastasis and response to treatment.
We are now in the age of big data in biology and medicine, and we certainly see regular use of genomics in the clinic, helping treating clinicians make decisions. However, to me, one of the major challenges is data integration across biological levels. How do the patterns of alternations we see at the DNA level translate to the cell level, then to the tissue, the organism, and end up being determinants of disease aggressiveness, patient response to treatment and prognosis? How do all these systems build up from the other? What are the overarching rules and patterns that govern these systems?
Can we, as mere humans, ever understand it? Or is it, as my undergraduate self once thought, just too complex?
I’ll let my postdoc self answer: No. We just need the right mindset, questions and tools. We just need computational biology.
Contact
- Email:
This email address is being protected from spambots. You need JavaScript enabled to view it. - Anna Trigos X
- Anna Trigos LinkedIn
Dr Dane Newman is a Postdoctoral Researcher in the Gene Regulation Laboratory headed by Professor Ricky Johnstone. His expertise includes T cell immunology and preclinical models of malignant and infectious disease.
Back in 1997, my unthinking 17-year-old younger self was obsessed with the ocean, fish, and fishing, and I vehemently wanted my future career to reflect this passion.

A favorite author of mine once wrote, “We spend most of our walking lives at work – in occupations often chosen by our unthinking younger selves.” Back in 1997, my unthinking 17-year-old younger self was obsessed with the ocean, fish, and fishing, and I vehemently wanted my future career to reflect this passion. Two years later, with high school complete, I left my childhood home in suburban Melbourne and journeyed off to Warrnambool on the south-west coast of Victoria to undertake a Bachelor of Science degree in Fisheries Management and Aquaculture at Deakin University.
Even though undergraduate coursework was largely uninspiring, it was my Honours year that gave me my first real taste of the scientific process, and I revelled in it! My mission that year was to investigate sea sweep – a poorly studied saltwater fish common along the southern Australian coastline and I spent a lot of time with a mask, snorkel and spear collecting specimens while being repeatedly battered by the rolling Southern Ocean swells. Although ocean diving was hard and not sustainable long-term, I relished the scientific process and took great joy and pride in piecing together the data and creating a coherent whole. And when I was offered a scholarship to undertake a PhD the following year, I jumped at the opportunity.
This time I chose to stay on dry land for my doctoral studies. My PhD research would investigate the reproductive biology and breeding of Murray cod (Australia’s largest freshwater fish species) within controlled indoor aquaculture systems. Data from my experiments resulted in the publication of four primary research papers and perhaps most importantly, my love of scientific research continued to grow. The science world had me hooked (excuse the pun), and with my PhD now accepted, I eagerly wanted to pursue this career path into the future.
But by now you must be wondering, ‘How the hell does a glorified fish farmer end up in cancer research?’ The answer is largely serendipitous.
The journey started only four months after my PhD graduation, when my life was completely upended. A routine blood test revealed a white blood cell count that was abnormally high. A bone marrow biopsy a week later confirmed a diagnosis of primary myelofibrosis – a chronic blood cancer that progressively scars the bone marrow ultimately leading to severe anaemia and a life expectancy of around 3-11 years. My only hope of a cure was a bone marrow transplant, but given my disease was still in its early stages, the transplant procedure carried too much risk to justify as a first line of treatment. I was told by my doctor that I would need to ‘earn’ my right to receive a transplant – that is, my body would need to deteriorate to a point where the risks of a transplant were no longer overshadowed by the immediate risks of the disease.
The poet Jason Shinder wrote that “Cancer is a tremendous opportunity to have your face pressed right up against the glass of your mortality.” At only 29 years of age, I suddenly found myself pinned up against that glass with no way of escape. Those first few months following the diagnosis were brutal and traumatic and any lingering thoughts about a professional career in science seemed no longer important, even trivial. Fortunately, I had my partner and future wife-to-be beside me, and she was (and continues to be) an incredible source of support and comfort. My medical journey also meant that I was back living in Melbourne and as time passed and the initial shock slowly dissipated, I started to find my feet again and began to look for work.
Unsurprisingly, there wasn’t a great deal of opportunities available for a newly graduated fish researcher in the big city, but I made the best of a bad situation. And after several months of submitting job applications, endless emails, phone calls, and door knocking, I was lucky enough to secure a short-term contract as a research assistant in Professor Graham Lieschke’s lab within the Cancer and Haematology Division at the Walter and Eliza Hall Institute (WEHI). A lot could be said about securing a post in a blood cancer research lab immediately after being diagnosed with blood cancer, but I can assure you that it was nothing but a coincidence - I certainly wasn’t harbouring any grand ambitions to find a cure for my own disease. I was hired because Prof. Lieschke’s speciality was zebrafish – a small ornamental fish species that is also an important biomedical animal model - and I was brought onboard to help create a library of cryopreserved zebrafish sperm that would act as a tissue bank for rare genetic mutants. The project was still a long way off conventional medical research, but the zebrafish job meant that I had my ‘foot in the door’ of the biomedical research community.
My time in the Lieschke lab only lasted 9 months before the research funds dried up and I found myself hunting for a job again. However, my ability to garner interest from potential employers felt a little easier this time around, no doubt supported by a sparkling reference from Prof. Lieschke and a modicum of new ‘biomedical experience’ listed on my CV. And soon enough I was offered a research assistant position within the Molecular Immunology Division at WEHI with Dr Erika Cretney and Prof. Gabrielle Belz. This time, there wasn’t a fish tank in sight, and I would now have to become accustomed to working with mice. Looks like I would now be staying on dry land for the near future.
The Belz lab focussed on a group of immune cells called ‘T’ cells. T cells are one of the body’s best defenses against infection and cancer and they represent a wonderfully fine-tuned killing machine. When T cells respond to an infection, they must orchestrate a difficult balance between efficiently killing infected/mutated cells, while at the same time minimising collateral damage to healthy tissues (and thereby avoiding autoimmune disease). A primary focus of the Belz lab was to identify the molecular machinery that would allow T cells to navigate this narrow bridge between health and disease. Although I found the subject deeply fascinating, my life in the Belz lab began with virtually no experience in cellular, molecular, or immune research, so my first couple of years were like learning a foreign language. Gratefully, I had some wonderful mentors in Dr Cretney and Dr Rhys Allan and over time, I gradually started to speak the language and find my groove.
After 5 years in WEHI’s Molecular Immunology Division, I had published a few immunology papers and I was offered my first postdoc position at the Peter Doherty Institute in the lab of Dr Laura Mackay and Prof. Frank Carbone. The focus in the Mackay/Carbone lab was on a specialised form of T cell called a ‘Tissue-Resident Memory T Cell’ or ‘TRM’ and I was there to help understand the unique molecular program that allows TRM to adapt and maintain permanent residence in non-lymphoid tissues such as the skin and gut. These investigations continued to expand my knowledge and insight into the immense plasticity and environmental adaption that T cells can exhibit both during and after infection.
During the latter part of my postdoc, 7 years after my initial diagnosis, my disease had started to catch up with me. A series of sub-standard liver function tests meant that the time had come to run the gauntlet and undergo a bone marrow transplant. Given my young age and relatively low number of co-morbidities, the odds of a successful transplant were in my favour. However, the risks of serious complications and death were still very real. One of the ways I dealt with the fear and uncertainty in the lead up to the procedure was to write about it. You can read about my transplant journey here.
My bone marrow transplant process began with a week-long intensive chemotherapy regime that destroyed my existing diseased bone marrow. Stem cells from a compatible donor (an anonymous gentleman from Queensland) were then infused into my bloodstream from which they migrated into the recently emptied cavities of my bone marrow. A couple of weeks later, the stem cells had regenerated into a brand new and healthy haematopoietic system. Pretty amazing hey! Of course, this description is an overt simplification of a complex procedure that carries many life-threatening risks and no guarantees of success. One of the major risk factors is graft versus host disease (GVHD), a situation whereby the newly developed immune system from the donor begins to recognise the host (for example, me) as foreign and begins to attack healthy tissues. About 10% of all transplant patients receiving stem cells from a donor will die from severe GVHD. The harmful effects of this disease are primarily orchestrated by killer T cells that have been let off the leash. Weak or imbalanced molecular signals that ordinarily instruct these cells to calm down, switch off, or die, mean that these cells are unable to curb their destructive enthusiasm and suddenly, the skin, liver and gut are no longer spared from immune attack.
In addition to GVHD, two other major risk factors following bone marrow transplantation are infection and relapse (that is, the original cancer returns). These situations often occur because of weak or suppressed immunity – essentially, the inverse of what occurs with GVHD. Here, the molecular machinery that enables T cells to activate, switch on and survive are imbalanced or malfunctioning, thus enabling opportunistic pathogens and malignant cells to grow and thrive. Therefore, for a bone marrow transplant to be successful and to avoid GVHD, infection and relapse, the newly established immune system must find and maintain a tightly regulated balance between hypo- and hyper-activation – the Goldilocks zone.
I owe my life to my donor’s perfectly balanced immune system. In the months after my transplant, I experienced a mild form of GVHD on my skin and liver, but it was never enough to cause any serious damage and it soon resolved. However, rather than being viewed as a burden, this mild and treatable form of GVHD was regarded by my doctors as an ‘encouraging sign’ and that my donor’s T cells were happy in their new home and functioning well. Not only would they protect me from infection, but any residual disease left in my system would be recognised and destroyed. Ultimately, the T cells from a stranger gave me a second chance at life and I’ll be forever indebted to him, as well as the medical community at Royal Melbourne Hospital and Peter Mac for giving me this extraordinary gift.
And while my T cells managed to destroy my disease, I also realise that a small molecular aberration here or there could have upset this delicate balance and left me in grave danger. It really is a fine line between cure and death when transplanting a new immune system.
These days, I work as a postdoc in the lab of Professor Ricky Johnstone at Peter Mac where we are investigating the use of small molecule inhibitors for reinvigorating a patient’s T cell response against tumours. We rarely notice when someone’s immune system is working against cancer, it is only in circumstances where tumours can escape immune control that we begin to understand the shortcomings of human immunity. In many cancer patients, the T cell response against the tumour has (over time) become hypo-activated as a protective mechanism to minimise collateral damage to healthy tissue. The problem is that the patient will die if the T cell response is not restored to a level where tumours can be cleared. My current research is trying to understand the molecular machinery that causes the T cells to suppress their activity and identify small pharmacological inhibitors that might help subvert these processes and re-establish the life-saving function of anti-tumour T cells. The balance must be restored!
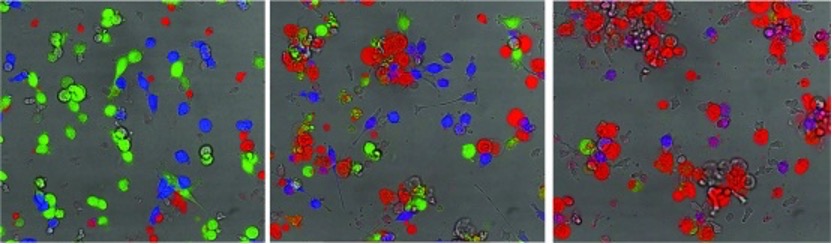
Figure legend: Time-lapse photography of anti-tumour T cells (small transparent cells) targeting and killing cancer. Live cancer cells are shown in green and turn red when destroyed by their killer. In contrast, another group of cancer cells (shown in blue) are unrecognisable to the T cells and largely escape death. Photo Credit: Kelly Ramsbottom.
Contact
Email:
Dr Jovana Maksimovic is a senior researcher in bioinformatics and computational biology in the Oshlack Lab at the Peter MacCallum Cancer Centre. Her work has primarily focused on epigenetics and transcriptomics data analysis and methods development projects, with a particular influence in the field of methylation array analysis. Jovana currently holds an NHMRC EL2 Investigator Fellowship and is the lead computational biology investigator on a Chan Zuckerberg Initiative Single-cell Biology Pediatric Network Grant.

I have always been interested in science, but my pathway to becoming a bioinformatician at Peter Mac was not linear and relied on more than a dash of serendipity.
My parents were both electronics engineers and my dad had a vast collection of “Galaksija” (translation: “Galaxy”) magazines - a popular science periodical published in Yugoslavia throughout the 70s and 80s - which I loved leafing through as a child. I was fascinated by everything to do with space and marvelled at the glorious, neon-infused illustrations of astronomical phenomena. I also became instantly fascinated by computers when dad brought home a ZX Spectrum computer that plugged into the TV.
We migrated to Australia in 1989 and I was absolutely overjoyed when we welcomed a Windows PC into the house five years later - enabling hours of computer tinkering! High school then inspired a love of biology; especially the bustling metropolis of molecular machinery inside our cells. But it was only when I picked up an unassuming cream-coloured pamphlet spruiking “bioinformatics'' at a university open day that I stumbled onto my future career. Bioinformatics uses computer science, mathematics and statistics to solve biological problems and it was a perfect fit for this computer-tinkering, biology-loving investigator!
After majoring in computer science, biology and genetics at university, my journey took a turn into agricultural research with the Victorian public service. There, I was involved in research focused on improving milk yield and quality from cattle using tammar wallabies as a model. This segued into a PhD investigating the expression and regulation of sialyltransferase genes in the mammary gland of marsupials and mammals. And, despite my love of computer programming and data analysis, it was an adventure to also work in the lab, and even the field - catching, handling and milking (!) wallabies.
My career then swerved into medical research at the Murdoch Children’s Research Institute in 2011. This coincided with the explosion in new massively parallel sequencing technologies, which revolutionised biology and medicine. As costs rapidly decreased and data generation exponentially increased, bioinformaticians like me became firmly entrenched at the frontlines of life sciences research, meaning that my work has touched everything from T-cell biology to the treatment of kidney disease.
By the time I arrived at Peter Mac in 2019 researchers were able to routinely sequence the DNA and RNA of millions of single cells, once again upping the ante for bioinformaticians. I am currently leading the computational analysis for the development of a globally-accessible cell “atlas” of the healthy paediatric airway, as well as a single-cell project focused on characterising inflammation in children with cystic fibrosis. This has led to an interest in developing better analysis methods and approaches for very noisy single-cell data specifically from clinical samples to help unlock their full potential in increasing our understanding of cells in health and diseases such as cancer. I am also very excited to be piloting an approach for detecting known cancer fusion transcripts by adapting a novel single-cell technology.
Contact:
Email:
Dr Katie Fennell is a postdoctoral researcher in the Eckersley-Maslin Lab. She has expertise in lineage tracing, single cell technologies and non-genetic evolution in cancer. Work from her PhD has been published in top tier scientific journals, including Nature and Nature Communications and she was a finalist for the prestigious Premier’s Award for Health and Medical Research in 2022. She is particularly interested in cellular behaviour and clonal biology and is currently investigating novel regulators of plasticity in cancer.
At the age of 10, I was convinced that I was going to be a forensic pathologist.

My love for science arose from an unusual and slightly morbid route. From a very young age, I was obsessed with crime shows such as CSI and Diagnosis Murder. The science of forensics fascinated me, but so did the problem solving required to crack an investigation. As a result, at the age of 10, I was convinced that I was going to be a forensic pathologist (I didn’t have many friends). However, once I started high school, I quickly realised that forensics wasn’t going to be half as glamorous as portrayed on TV. Instead, I channelled this early interest in science and curiosity into studying human biology.
At school, I really loved the practical aspect of science where my skills shone. My biology teacher once told me: “Your coursework and essays are atrocious, but your lab work is excellent.” This was (unfortunately) true, and it resulted in my first failure as a young scientist whilst studying for my A levels (the UK equivalent of Year 12). Based on my predicted grades, I received an offer to undertake a Bachelor of Biomedical Science at the prestigious King’s College London. Much to mine and my parents’ horror, my terrible essay writing skills led to me flunking my exams, and I ended up going to a mediocre university located in Bristol. Luckily for me, I loved living in Bristol, and I was thankful for this early failure. It made me realise that I needed to hone my scientific skills outside the lab if I wanted to have a successful career in science.
Again, at university, I felt so natural being in lab practicals. However, I found sitting in lecture theatres and attempting to memorise endless signalling pathways slightly dull. I just wanted to put on a lab coat and start doing experiments. This theme had been so pervasive throughout my studies, so I decided to pursue a career in laboratory research following my undergraduate degree. I also decided to do this on the other side of the world. So, at 21, I packed my bags and left the small English town where I had grown up and moved to Melbourne, Australia. At this stage, I was particularly interested in how the brain worked and so I began an internship in the Neural Plasticity and Epigenetics lab at the Florey Institute of Neuroscience. This is where things finally started to make sense for me as a scientist. It wasn’t exactly a lightbulb moment, perhaps more of a slow burning candle, but the barrier that I’d experienced between scientific theory and the work I was conducting in the laboratory had finally started to dissolve.
My interest in epigenetics really took hold at my time at the Florey and I wanted to focus on this for my PhD, so I joined the Cancer Epigenetics lab led by Prof. Mark Dawson in 2016. Until this day, I am not sure why Mark took me on as a student as I had limited laboratory skills, I only had experience in qPCR and mouse brain dissections. He also gave me 2 extremely high-risk projects. The first project was optimising plate based single cell RNA-sequencing to study tumour heterogeneity in patients with leukaemia. The second was to develop a novel single cell “barcoding” technology that would allow the tracking of individual cancer cells over time. At the time, single cell technologies were in their infancy, and I really felt like I was in over my head. Embarking on these ambitious projects is where I experienced my next set of failures, following my exam blunder from all those years earlier.
For the first 2 years of my PhD, nothing I did in the lab worked. I barely had any useful data, and I was taking 2 steps forward and 1 step back. I tried to stay positive, but some weeks were bleak. As laboratory scientists, we are so intimately and personally tied to our projects, so it can be incredibly hard not to place blame on yourself when things aren’t working. During this tough period, I had to keep reminding myself that if the technology we were trying to develop was easy, then someone else would have done it already. I somehow found the resilience to persevere and with the help and guidance of many colleagues and friends, things started to finally work. It was amazing how quickly our story came together once the barcoding technology had been optimised. In contrast to how down I was in the early stages of my PhD when things weren’t working, this latter stage of my PhD was some of the most exciting and rewarding phases of my career thus far. Research is a rollercoaster ride, and you must experience the low points to appreciate the amazing highs.
I’ve now started my postdoc with Dr. Melanie Eckersley-Maslin and I am building upon some of the themes from my PhD in the exciting area of cancer cell plasticity and non-genetic cancer evolution. For decades, the field of cancer research has focused on mutations involving the DNA sequence and how these are crucial for cancer development. However, we now realise that non-genetic factors, such as the transcriptional and epigenetic landscape of cancer cells, might be equally as important in driving tumour evolution. I am incredibly lucky to be working in this exciting area and am hopeful that my research can add a small piece to the ever-growing puzzle of cancer biology. I won’t lie- some days, I still feel like that naive girl who could only do qPCR and I still fear that my experiments won’t work. I have come such a long way since I started my PhD and each failure, I have experienced has taught me something valuable.
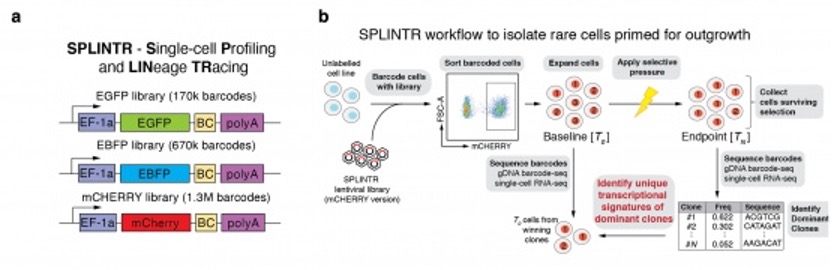
Figure Legend: Single-cell Profiling and Lineage Tracing with expressed barcodes (SPLINTR)
- Three independent SPLINTR barcode libraries were generated, each containing a distinct fluorescent reporter and barcode structure. SPLINTR barcodes are constitutively transcribed upon genome integration, enabling tracking of clonally related cells together with their individual transcriptomes.
- Schematic detailing the experimental design used in SPLINTR experiments to retrospectively identify the transcriptional signatures of dominant clones. This technology was developed as part of my PhD project.
Contact
Email:
Dr Kazuhide Shaun Okuda is a Postdoctoral Fellow in the Hogan laboratory, which is part of the Organogenesis and Cancer Program within the Peter Mac Research Faculty. His expertise includes vascular biology, zebrafish research, disease modelling, imaging, and drug discovery.

Whenever my mother took me to morning wet markets in Malaysia, I would go straight to the fish vendor and sit there looking at all sorts of different fish. I always wanted a fish, but my mother would stop me, as I didn’t have a particularly good track record of keeping pets. Now, I am surrounded by thousands of fish as I use the zebrafish model to study how blood and lymphatic vessels develop. Zebrafish, a popular pet fish, is an excellent animal model for biomedical research as it has a highly conserved genetic control of development and disease. Importantly, by fluorescently tagging specific organs of the fish, we can follow how these organs develop in real-time because zebrafish embryos are transparent during development and develop rapidly. As human cancer cells can be transplanted to both zebrafish embryos and adults, we can also model specific cancer cell behaviours, observe the molecular drivers of metastasis, and test the efficacy of new cancer drugs. As a vascular biologist, I use zebrafish with fluorescent blood and/or lymphatic vessels and observe how they grow and respond in diseased conditions. I can spend hours looking at these colourful vessels under a microscope without getting bored because they are so beautiful! It is important to study how blood and lymphatic vessels develop as they play roles in various human pathologies such as cancer, chronic inflammation, and organ transplant rejection.
My main research focus is to generate and characterise genetically modified zebrafish (zebrafish transgenics) that enable visualisation of the dynamic activity of key signalling pathways for blood/lymphatic development in real time. Apart from development, we are interested in utilising these zebrafish transgenic lines to understand molecular mechanisms of blood/lymphatic vessels in various pathological settings in real time. Apart from this, I am interested in uncovering new lymphatic genes that may have functions in both developmental and pathological lymphangiogenesis. We are hoping to utilise the information we gain from this research to identify novel therapeutic targets/agents for cancer. One discovery that I am particularly proud of is when I contributed to the generation of the first lymphatic “atlas” that described all the lymphatic vessels in zebrafish larvae. This was made possible due to us generating the most lymphatic-specific fluorescent transgenic line in zebrafish at the time. This allowed us to visualise previously hard-to-see lymphatic vessels in the face and the intestine of the zebrafish and I even had the privilege to name some of these vessels. Using this information, we demonstrated that lymphatic vessels in fish respond to inflammatory stimuli from immune cells and revealed that a subset of lymphatic vessels originate from a non-venous origin, something that has never been live imaged before. Many researchers in the field have also used the tools and information we generated to:
- identify various new genes essential for lymphatic development,
- show that lymphatic vessels in the heart contributes to cardiac regeneration in fish, and
- identify a novel mural endothelial cell population that may contribute to clearance of brain waste and brain vascular regeneration.
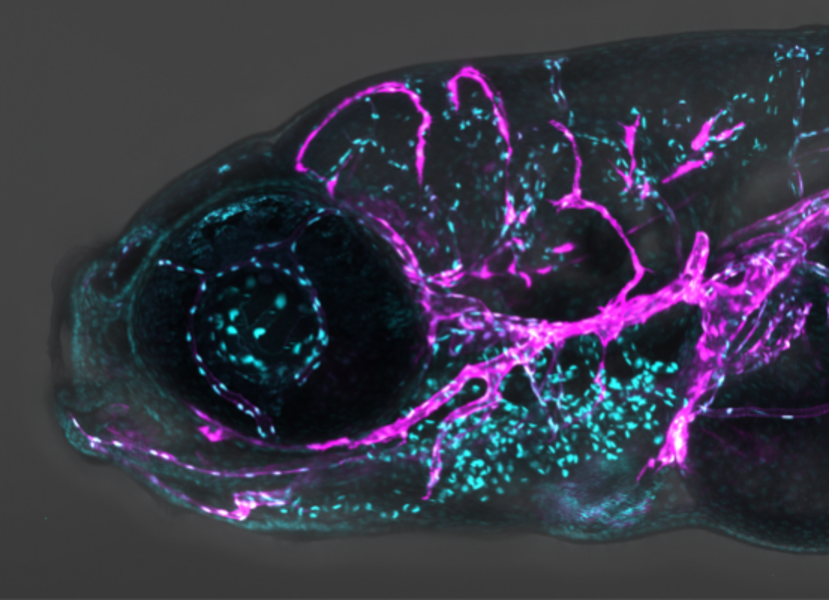
Figure legend: The beautiful facial vessels of zebrafish. Cyan: Endothelial cell nucleus Magenta: Lymphatics and vein
The angiogenesis and lymphangiogenesis field are rapidly growing and so, it is essential for me to be in an environment that enables me to continue doing innovative science. I am therefore excited to be part of the Peter MacCallum Cancer Centre, which is in the heart of the biomedical Parkville precinct, with excellent world-class facilities. Here at Peter Mac, I am working side by side with scientists making an actual impact on the lives of cancer patients. Also, I see cancer patients every day when I come to work reminding me of how important it is to progress our understanding of human disease via research. I see research as a huge team effort. I believe that whatever small discoveries we make regardless of it being basic or translational, will one day collectively contribute to treatments for human patients. This motivates me to strive through the harsh environment of academia. I hope one day, the tools and information I generated would contribute to the betterment of human health and zebrafish would be world famous!
Contact
- Email at
This email address is being protected from spambots. You need JavaScript enabled to view it. - Dr Kazuhide Shaun Okuda X
- Google scholar
- Publons
Mark Li is a Research Officer at The Victorian Centre for Functional Genomics. He is specialised in assay development and drug screening, high content imaging in 3D organoid models, high throughput transcriptomics, and machine learning based analytics.
After a boring semester as an electrical and computer engineering student, I quit university and moved to Australia in 2009 with no connections – or really any idea of where the country was. My parents called me mad, and if you grew up in China back then, you would understand. In retrospect, they were probably right (considering most of my classmates are now at Alibaba or Tencent-like companies). But they did respect my decisions. Interested in medicine and biology, I enrolled at the newly formed Department of Biomedicine at The University of Melbourne. While most students were focused on getting into medical school, I chose the research track, majoring in immunology with a minor in economics and finance. I would probably say that my love for research was at its purest during that time. As a broke undergraduate student, I let go of a job at a venture capital firm to study immunology. The idea of an active, trainable system already built into our bodies and able to respond (or not) to future threats (pretty much anything) was, and still is, fascinating to me.
Knowing virtually nothing about careers in medical research, and with few peers around me, I turned to my cousin (who was then a postdoc at Genentech) for consultation. If I took any lesson away as a hot-headed 20-year-old who reckoned the sky's the limit, it was that I need a better platform. I was fortunate enough to take on my Honours project at WEHI, a fertile soil for learning.
My Honours year turned out to be unexpectedly challenging. The transition was difficult, and it was hard to communicate with my direct supervisor Finding balance also proved to be difficult. Doubling down on hard work was not the solution to all problems (as in the real world, Martingale processes don't always work). However, it was a year of growth where I learned from trial and error, and recognised what I needed to do next. Luckily, a great supervisor and mentor came along at the right place and time. Although the field of research was drastically different to my Honours year, I still managed a successful and enjoyable PhD journey. If you are interested, you can find my publications on killer protein BAK and VDAC2 on mitochondria regularions through a quick Google search. The main idea is to position yourself well and embrace change without fear.
At the end of my PhD and thanks to a generous WEHI scholarship, I was able to travel to America, Europe, and Japan for meetings and to discover what I wanted next. There were quite a few offers on the table and a question remained unanswered: "Why more cell death?" I knew I was supposed to build a "track record" for my fellowship, but I was not sure if I would be happy to drink the same bottle of wine, more or less. I wondered whether time spent in a similar field would generate enough returns – at least measured by felt happiness. Soon after one too many beers, a soccer injury, and swimming (not in that order), I found myself running late to my poster presentation at Cold Spring Harbor meeting on Cell Death. There, I bumped into Professor David Andrews (director at Sunnybrook Research Institute in Canada), who challenged me to do my project in a faster, better way. Later in the year, after a follow-up meeting with him at a cafe table in Melbourne’s Parkville, I decided to take a chance on Canada. After all, he was willing to take a bet on me with a big project (I knew nothing about imaging, microscopy or robotics at that time, let alone, machine learning; the best code I wrote was "hello world"). And, what was there to lose?
The perk of being a postdoc is being able to face a series of challenges and solve them, while accumulating incredibly valuable career and life experience. David will never hold your hand, but he will resource a project with whatever you need to learn and get things done. I am grateful for my first postdoc experience. I have gone through the thrill and stagnation of starting up a biotech company from the ground up, dabbled with consulting, learned a basket of skills in precision medicine, and hopefully have a good paper coming out soon.
But that is not "beyond" enough, is it? Thanks to the skills I developed during the postdoc phase, and more importantly, the friendships (majority non-scientists) that inspired me to stay curious, I was fortunate and bold enough to self-introduce myself to the founder of a fintech company. I did not meet any of these inspiring people at networking events. Just old school emailing. Be sincere, be honest. This has been a very interesting period of my life, and there is not doublt I will benefit from the knowledge and ongoing relationships. Apart from luck, I can only say, "Please be bold, be curious, and have a go.”
Time to land back on today. Biomedical research still has a special place in my heart. Giving it a little historical perspective, biotech had its boom after the tech boom 20 years ago (and the unfortunate crash, partially due to the failure to scale). We currently have all these amazing technologies thanks to advancements in computational power and some aspects of applied physics. My skill set is shaped to provide platform technologies that enable researchers to do more at scale. There's no better place to do this than the Victorian Centre for Functional Genomics in Australia, which has a steady supply of biology (academic or commercial, basic or translational), good infrastructure, and most importantly, the vision and agility to change.
I wouldn't know what to answer if you asked me "So, what's next?" I have some vague ideas, but I don't know what to call them yet. I never really liked the usual, and my untested ideas might well someday blow up in my face. Fail well, spectacularly. Or succeed, equally well.
Dr Mirren Charnley is Senior Research Fellow at Peter Mac.
She works within the in the Russell laboratory, which is part of the Research Faculty’s Organogenesis and Cancer Program.
Her expertise includes developing T-cells, biomaterials, microscopy and flow cytometry.

Someone recently asked me if I wanted to be a scientist when I was a child.
The honest answer was: not really.
Not because I don’t love science, which I do, but because when I was young I didn’t really know what a scientist was.
However, I always enjoyed science, it made sense to me or felt like my language.
So, I went to the University of Sheffield for my undergraduate degree, combining molecular biology and chemistry.
Through this I learned that I love both areas, but am terrible at the practical side of organic chemistry. I spent four years trying to make fine white powders and getting dirty yellow oils.
I decided life as an organic chemist was not for me, so I instead completed a PhD in cell biology and biomaterials. I have been, more or less, at the interface of these two areas ever since.
For my first Postdoctoral appointment, I veered towards the biomaterials side. I headed to ETH Zurich, a public research university in Switzerland. My role was as a cell biologist who was going to guide biology-related projects within a polymer chemistry group.
There, I spent my time making cell-sized microwells in various shapes and with a range of coatings. I used these to explore how certain cues and 3D cell shapes drives cell behaviour. I particularly liked watching the cells divide but I wanted to go deeper into the biology that drives the process, so I decided to veer back towards the biology side of my research.
At this stage my partner and I decided to leave Switzerland. We hadn’t been that successful at learning German and we decided to try life in an English-speaking country.
This is how I embarked on a post-Doctoral position with Professor Sarah Russell. With Sarah, I explore how T cell development occurs, and how polarity and division influences cell fate.
I hope this knowledge will eventually help to provide better immunotherapies or immunomodulatory treatments.
I think one of the hardest things for me is choosing the best time to become a Group Leader. I am still a Senior Research Fellow, partly because I love playing in the lab too much, puppeteering my cells and watching them divide. I do feel a need to justify this choice because at times it’s hard to shake off the feeling that you have failed as an academic. Irrespective, I still love being a Senior Research Fellow and I am off to the lab to watch my pretties divide.
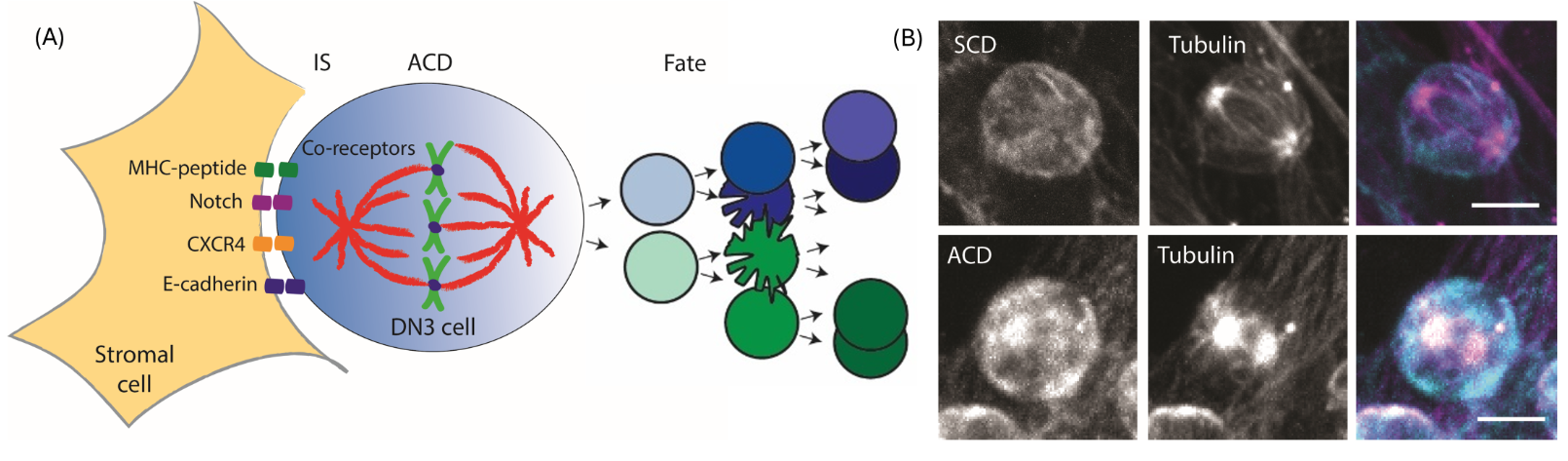
Figure legend
(A) I study how cues within the cellular environment steers the formation of an immunological synapse (IS) and how the cell divides to control cell fate during a critical stage of T cell development called β-selection.
(B) Representative projected z stack images of a dividing cell undergoing symmetric cell division (SCD) and asymmetric cell division (ACD). Bar = 5 µm.

